-
Overview of AlloReverse
-
Software Requirements
-
Tutorials
-
Example 1
-
Example 2
-
Input data format
-
Output result explain
-
Job Queue
-
Method
-
Overview of AlloReverse
Allostery is the phenomenon that functional site on protein is regulated by a topologically distinct allosteric site. Designing drugs binding this site could achieve better safety and selectivity. However, lack of global allosteric network throughout macromolecules is prohibiting our way forward.
AlloReverse aims to provide multi-scale information of allosteric regulation based on "reversed allosteric communication" theory, which indicates that allosteric sites are also regulated by orthosteric site. The reversed nature of the theory has provided possibilities for synchronous analysis of different allosteric regulations. AlloReverse could predict allosteric residues, allosteric sites, regulation pathways and correlation among allosteric sites. Especially, our server focused on hierarchical relationship among different regulations, and could offer a whole map of allostery on protein. Therefore, AlloReverse is believed to aid target identification, drug design and biological mechanism understanding.

-
Software requirement
The AlloReverse requires a modern web browser with JavaScript enabled. To view the submission status of a job, pop-ups must not be blocked. The following browsers have been thoroughly tested with AlloReverse:
Mozilla Firefox, version 5 or above(Download);
Chrome, version 9 or above(Download);
Safari, version 5 or above(Download);
Edge, version X or above(Download);
The latest version of Firefox and Chrome is recommended for visualization.
Below are the detailed operation system and brower version we have tested that our website can opertate successfully.
OS Version Chrome Firefox Microsoft Edge Safari Windows 10 109.0.5414.120 109.0 109.0.1518.78 not tested Windows 10 99.0.4844.51 109.0 109.0.1518.78 not tested Windows 10 98.0.4758.82 109.0 109.0.1518.70 not tested Windows 7 108.0.5359.125 103.0.1 109.0.1518.78 not tested MacOS Monterey 109.0.5414.119 109.0.1 109.0.1518.78 15.1 (17612.2.9.1.20) Linux CentOS 109.0.5414.119 n/a 107.0.1418.52 not tested Linux Ubuntu n/a n/a n/a n/a -
Tutorials
We would introduce how to use and how to analyze AlloReverse with two examples below.
Tutorials Table Example Name Example Protein Upload File Run Time Tutorial Example 1 HRas PDB ID: 4DLR Less than 1 minute View Example 2 SIRT3 PDB ID: 4FVT Less than 1 minute View -
Example 1
Introduction
HRas is a GTPase. It is also a vital protein in cancer development. Many efforts have been done on H-Ras. An allosteric site has already been reported on H-Ras (4DLR). Here, we would attempt to study possible allosteric regulations on H-Ras with AlloReverse.
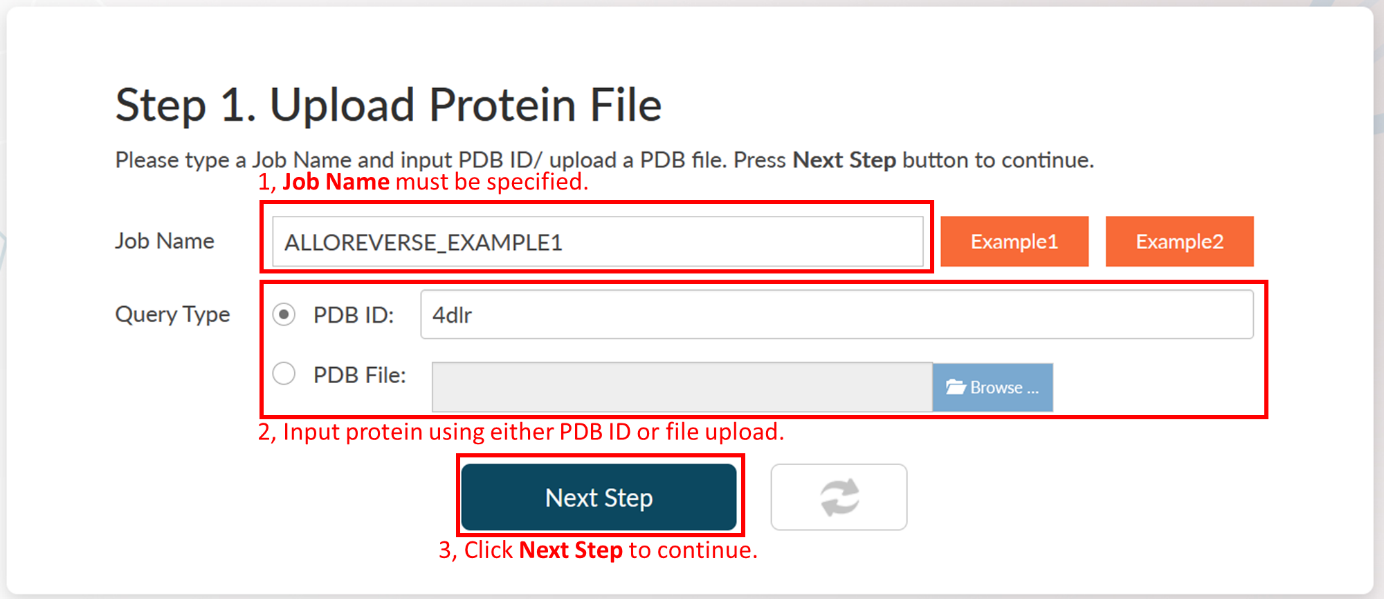
Step 1: Upload A PDB With Orthosteric Ligand.
AlloReverse requires two inputs: Job Name and a Protein File. The Job Name could be any string. The Protein File should be an interested protein binding orthosteric (or functional) ligand. It could either be a PDB ID or a PDB format file uploaded by users. More information about Protein File could be found in Input data format in the help document.
Click Home of AlloReverse menu, page down and find the Job Submit form. Press the Example1 button. AlloReverse will automatically load parameters for the user in the Job Submit form.
1. Input the Job Name "ALLOREVERSE_EXAMPLE1".
2. Select PDB ID as the query (input) type.
3. Input the PDB ID "4DLR", which is the structure of HRas binding GNP in orthosteric site.
Click Next Step to continue.
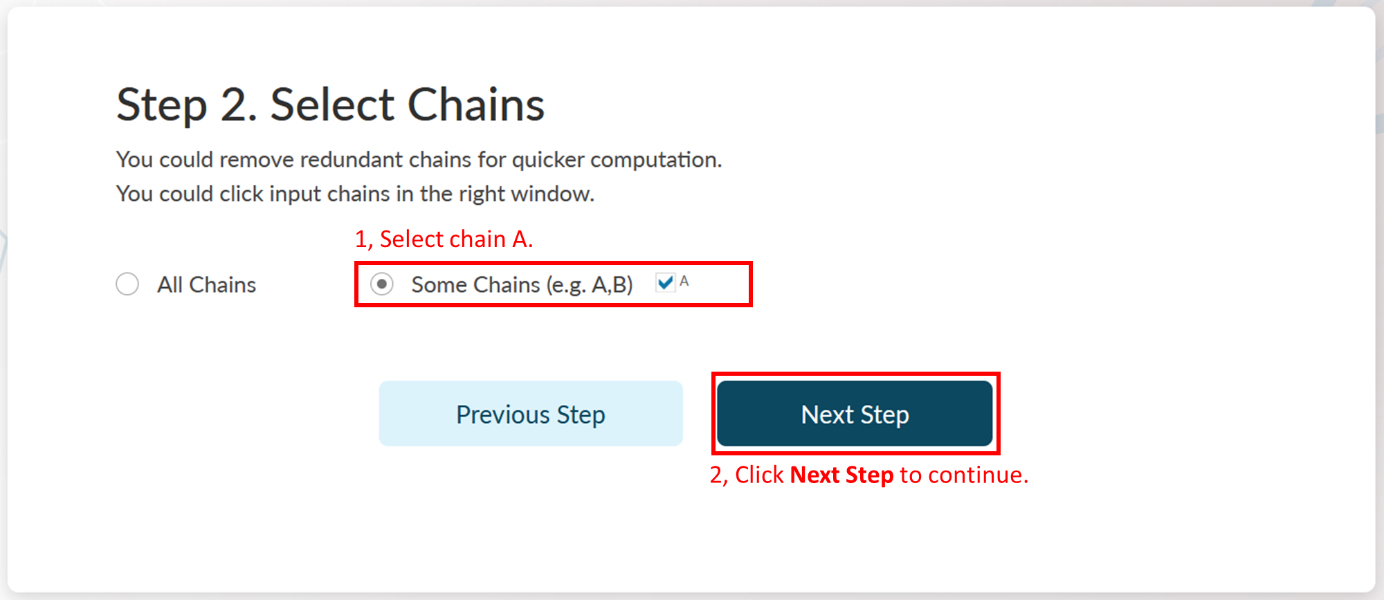
Step 2: Select Chains.
A PDB file could be a multimer containing many repeated amino acid chains. If the allosteric regulation across multiple chains is not considered, it is recommended to select only one chain for prediction. Because the more chains, the slower the analysis is. Note that if the orthosteric ligand does not have the same chain ID with the protein (e.g., the ligand is a peptide), you should also select it. Chain Indexes will be resolved by AlloReverse and users could select their interested chains in Step 2.
In Step 2 of Example 1, it could be found that there is only one chain (A) in PDB file "4DLR". The chain "A" is automatically selected by AlloReverse. Click Next Step to continue.
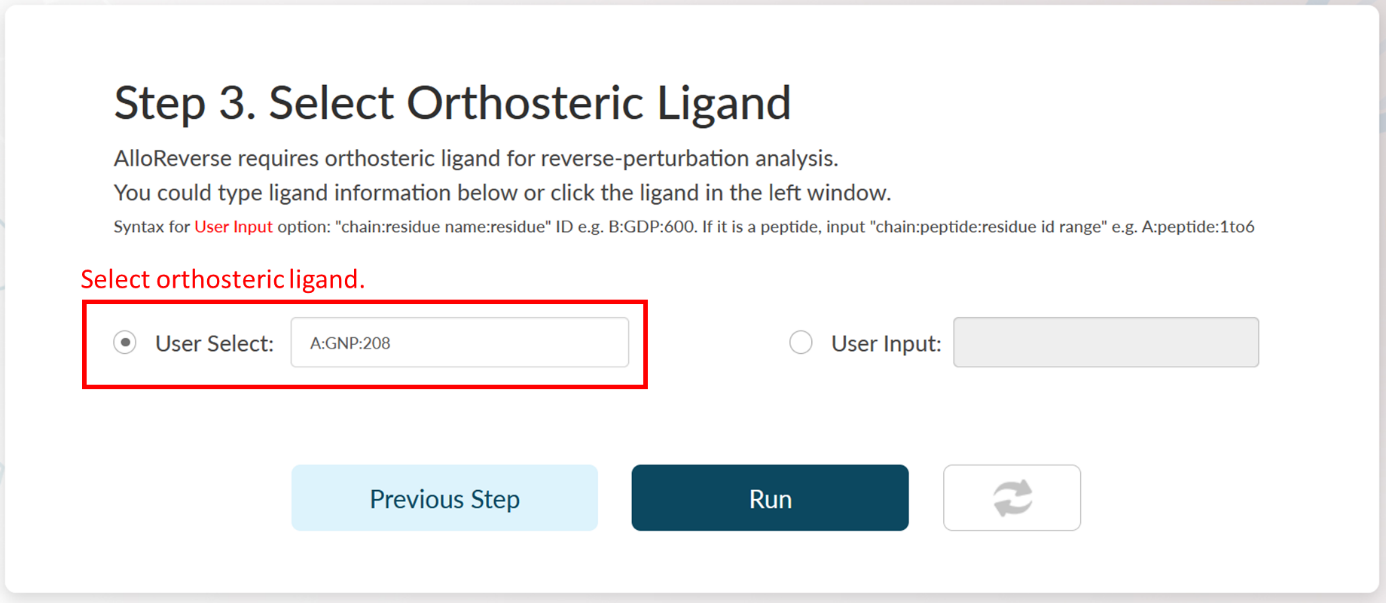
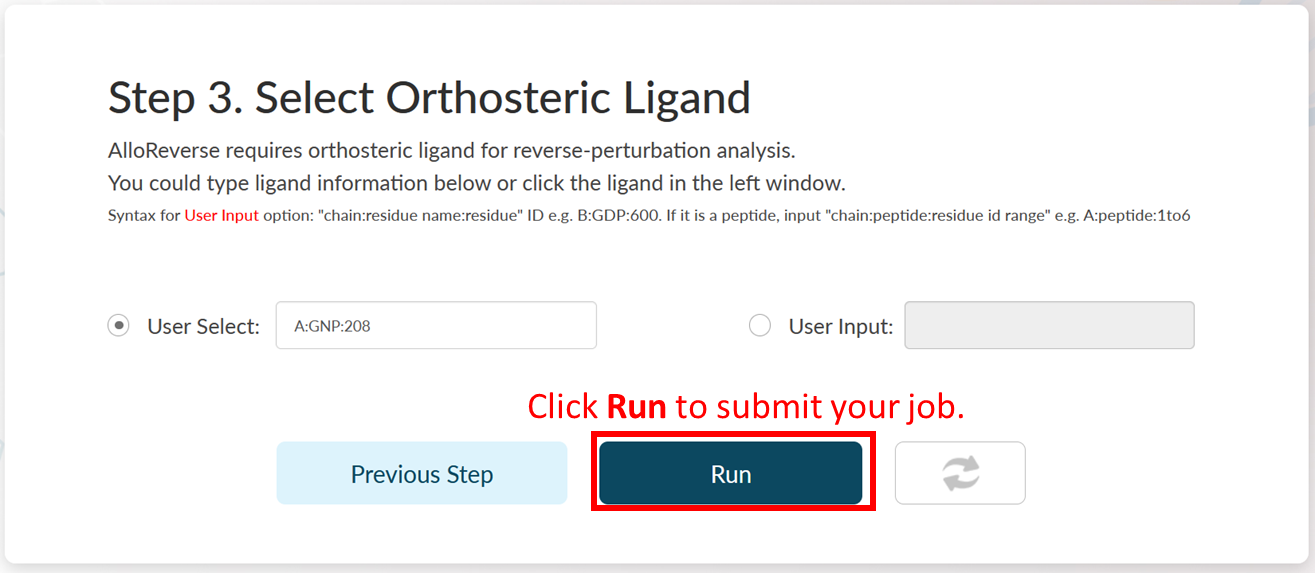
Step 3: Select Orthosteric Ligand.
AlloReverse studies allosteric regulations based on reversed allosteric communication. Therefore, the orthosteric ligand should be defined. AlloReverse would resolve all ligands in selected chain, so that users could pick one of them as the orthosteric ligand in the left choice box. If the orthosteric ligand is not labeled as "HETATM" in PDB file, users could also manually input the ligand int the right input box, in a format of "chain:residue name:residue id" (e.g., B:GTP:336). If the orthosteric ligand is a peptide, a manual input is also required in the format of "chain:peptide:residue id range" (e.g., C:peptide:1to6).
In Step 3 of Example 1, A:GNP:208 is the orthosteric ligand. Here, the ligand is automatically selected by AlloReverse.
Job Submission
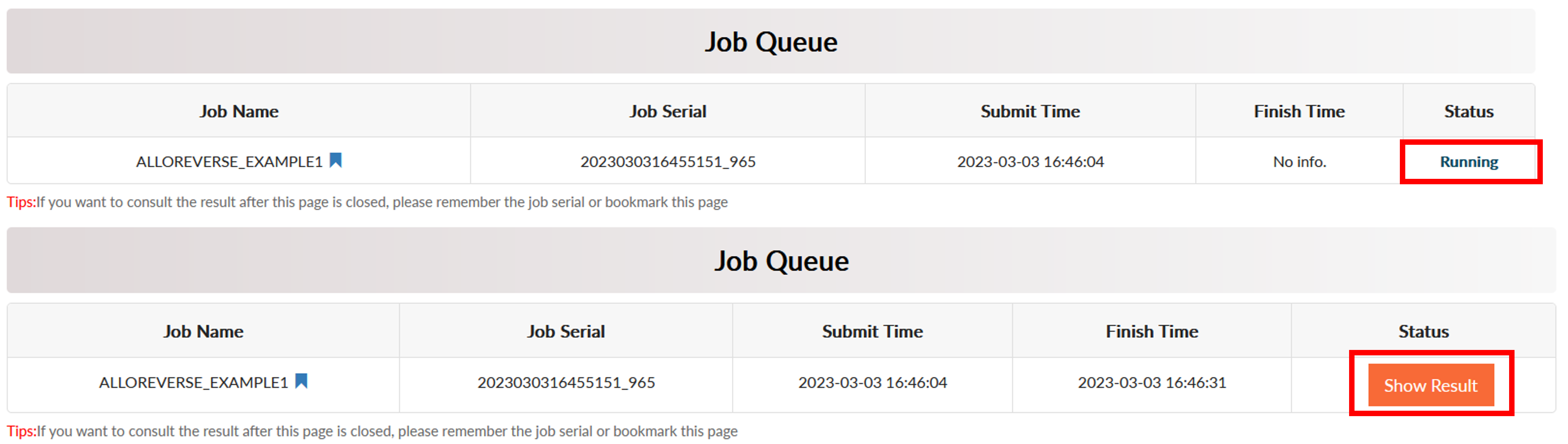
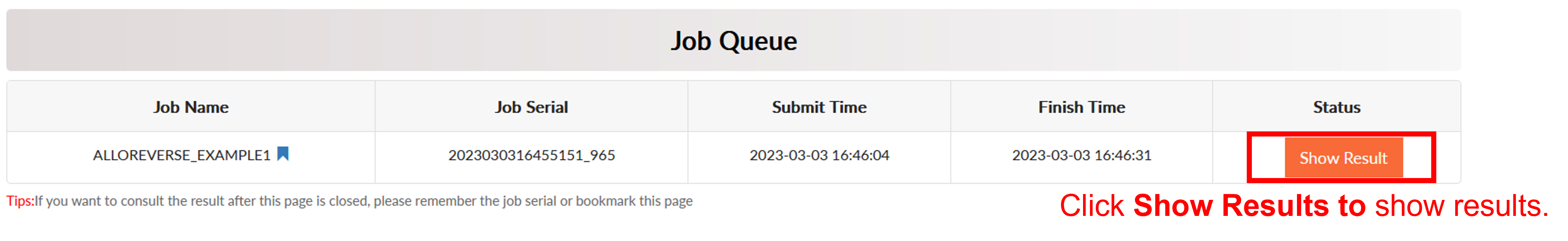
Click Run to start prediction and the job will start to run. The server would jump to the Job Queue page. It usually takes less than 1 minute for multi-scale analysis of allosteric regulations. When the analysis is over, the Status would become Show Result.
AlloReverse Result
Click Finish for reading prediction result. The result page contains four main sections: PDB Display Tool (top left), Job Information (top right), PDB Display Control Panel (middle right) and Result (bottom). The Result sections is be split into 2 subsections. The first subsection is about predicted allosteric sites with functional residues in it. Predicted sites are ordered according to prediction confidence. In Example 1, 2 allosteric sites are predicted, where Site 2 is a reported site and Site 1 is a novel site.

Users could click the Show Site to view the allosteric site in PDB Display Tool.
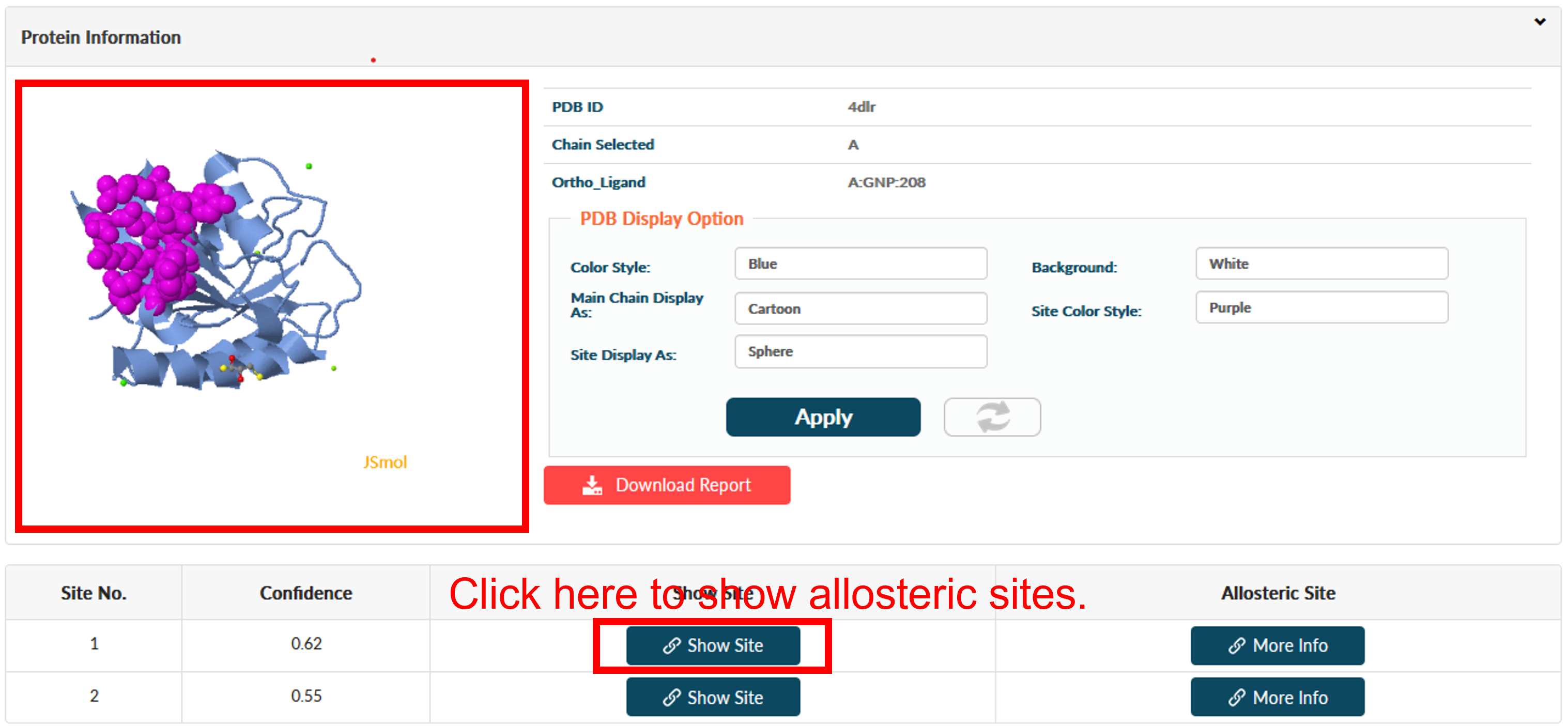
Users could click More Info for extended information about this site. Three parts of information are supported.
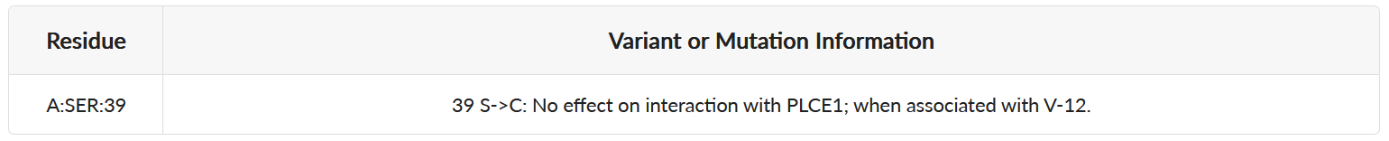
(1) Basic properties of the predicted pockets. This part includes volume, solvent-accessible surface area (SASA), and reversed allosteric effect (RAE) and residues in the site. Volume and SASA suggests the druggability of the pocket. RAE, or response against allosteric perturbation, is calculated by summing the RAEs of each residue in site. Site with higher RAE suggests stronger ability to regulate protein function.
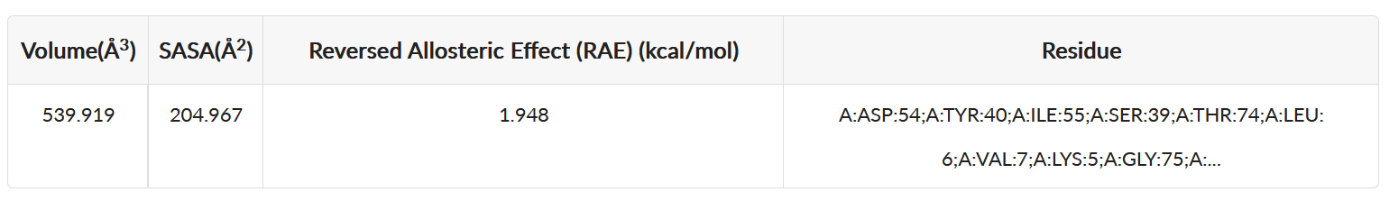
(2) Functional Residues in Pocket. Key hydrophilic and hydrophobic residues. Residues in sites are plotted based on their response against orthosteric perturbation (reversed allosteric effect, RAE) and hydrophilia. Residues with high RAE are considered to have key functions. In Example 1, it could be viewed that Tyr 71 might be important hydrophobic residues (hydrophilia < 0) while Thr 74 might be vital hydrophilic residues (hydrophilia > 0). This information could help further structure-based drug design and optimization on this site.
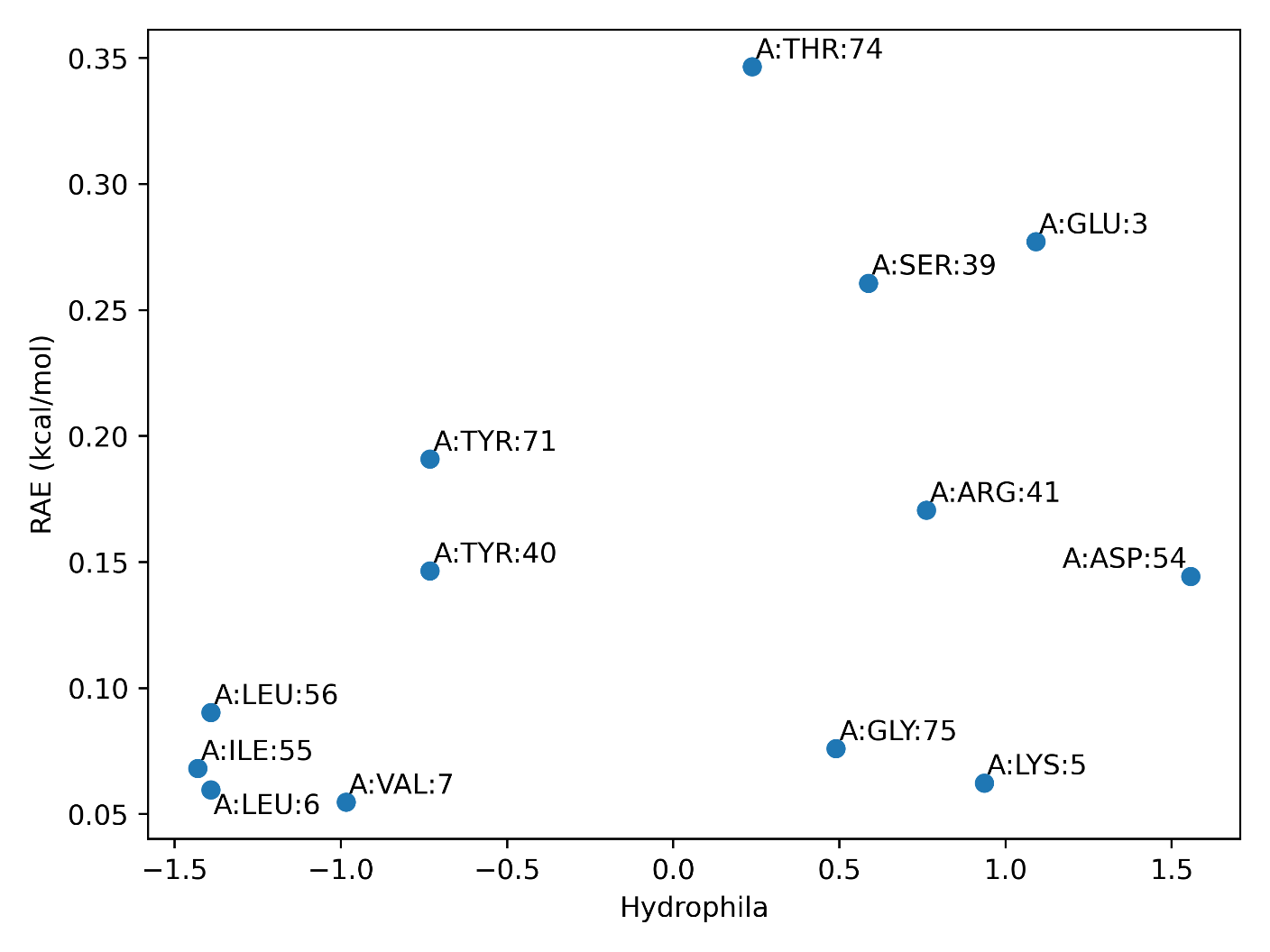
(3) Variants or Mutations. Variants and mutations recorded in Uniprot is mapped here to suggest possible functional or less functional residues in the predicted allosteric site. In Example 1, Ser 39 is recorded to have a nonsense mutation to Cys, suggesting its weak function in allosteric communication towards PLCE1.Note that this analysis is only available when PDB ID is used as input.
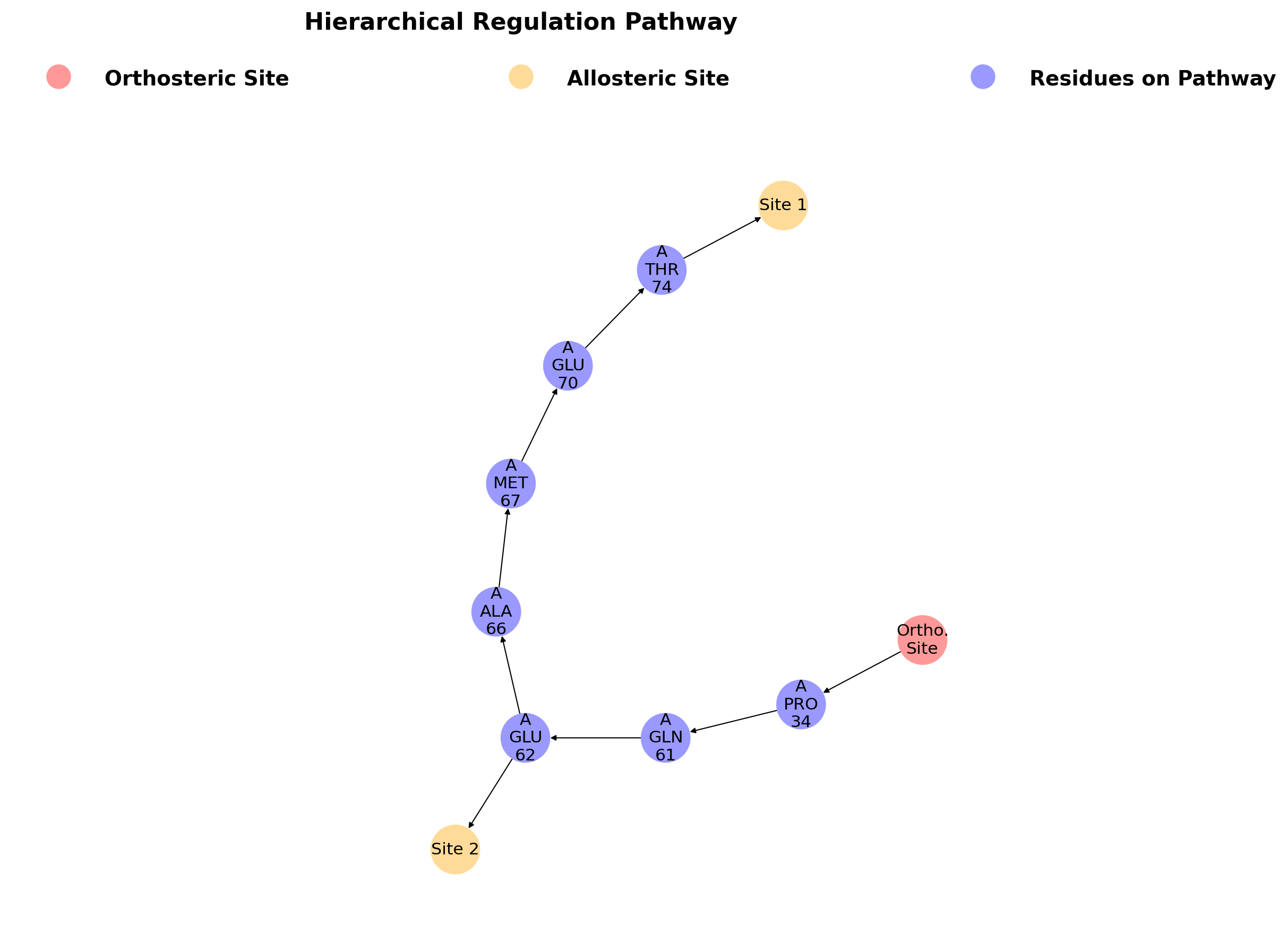
The second subsection focuses on hierarchical regulation pathways in protein, calculated from the orthosteric ligand to the most perturbated residue in site. Different allosteric regulation pathways could have couplings. A main feature of AlloReverse is to analyze the relationships among allosteric regulations and allosteric sites, offering a whole map of allostery. In HRas, it could been seen that Site 1 and Site 2 share common residues (Pro 34, Gln 61 and Glu 62) and regulation pathway.
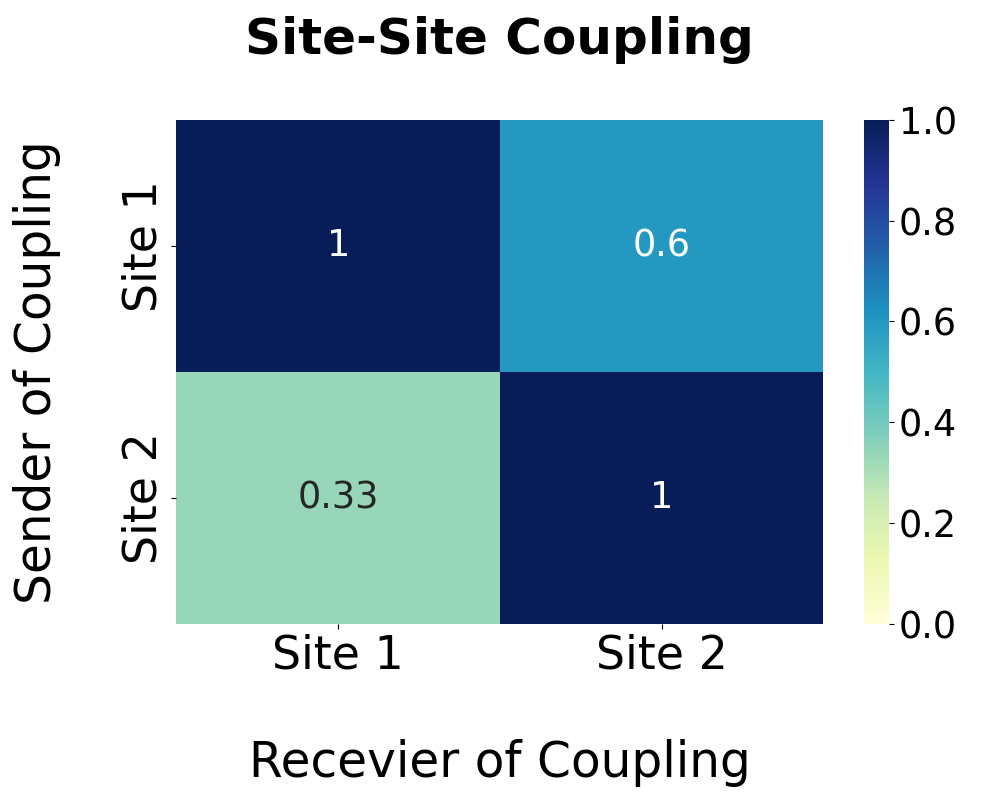
Based on the pathway analysis, regulations among predicted allosteric sites are further calculated and shown in the following heatmap. The degree of regulation of site A by site B is calculated by the proportion of pathway residues of site A shared by site B. Note that the site-site regulation analysis is only available when more than one allosteric site is predicted.
Finally, all information could be downloaded from the website. The package mainly contains a CSV containing up mentioned data and a PSE file (open by Pymol) for visualizing predicted sites.
-
Example 2
Introduction
Sirtuins 3 (SIRT3) is a deacetylase in mitochondria It plays vital roles in metabolic disease and senility. Previous studies have found 2 possible allosteric regulators on SIRT3 (PDB: 4C78 and 5Y4H), but they both bind at shallow pockets. In this example, we will try to study the overall allosteric network on SIRT3 with AlloReverse.
Step 1: Upload A PDB With Orthosteric Ligand.
AlloReverse requires two inputs: Job Name and a Protein File. The Job Name could be any string. The Protein File should be an interested protein binding orthosteric (or functional) ligand. It could either be a PDB ID or a PDB format file uploaded by users. More information about Protein File could be found in Input data format in the help document.
Click Home of AlloReverse menu, page down and find the Job Submit form. Press the Example1 button. AlloReverse will automatically load parameters for the user in the Job Submit form.
1. Input the Job Name "ALLOREVERSE_EXAMPLE2".
2. Select PDB ID as the query (input) type.
3. Input the PDB ID "4FVT", which is the structure of SIRT3 binding CNA in orthosteric site.
Click Next Step to continue.
Step 2: Select Chains.
A PDB file could be a multimer containing many repeated amino acid chains. If the allosteric regulation across multiple chains is not considered, it is recommended to select only one chain for prediction. Because the more chains, the slower the analysis is. Note that if the orthosteric ligand does not have the same chain ID with the protein (e.g., the ligand is a peptide), you should also select it. Chain Indexes will be resolved by AlloReverse and users could select their interested chains in Step 2.
In Step 2 of Example 2, it could be found that there are two chains (A and B) in PDB file "4FVT". We would only adapt "chain A" for further prediction. Here, "chain A" is automatically selected by AlloReverse. Click Next Step to continue.
Step 3: Select Orthosteric Ligand.
AlloReverse studies allosteric regulations based on reversed allosteric communication. Therefore, the orthosteric ligand should be defined. AlloReverse would resolve all ligands in selected chain, so that users could pick one of them as the orthosteric ligand in the left choice box. If the orthosteric ligand is not labeled as "HETATM" in PDB file, users could also manually input the ligand int the right input box, in a format of "chain:residue name:residue id" (e.g., B:GTP:336). If the orthosteric ligand is a peptide, a manual input is also required in the format of "chain:peptide:residue id range" (e.g., C:peptide:1to6).
In Step 3 of Example 2, seven ligands are resolved in the left, including A:CNA:401, A:SO4:402, A:SO4:403, A:SO4:404, A:ZN:405 and A:GOL:406, while A:CNA:401 is the orthosteric ligand. Here, the ligand is automatically selected by AlloReverse.
Job Submission
Click Run to start prediction and the job will start to run. The server would jump to the Job Queue page. It usually takes less than 1 minute for multi-scale analysis of allosteric regulations. When the prediction is over, the Status would become Show Result.
AlloReverse Result
Click Finish for reading prediction result. The result page contains four main sections: PDB Display Tool (top left), Job Information (top right), PDB Display Control Panel (middle right) and Result (bottom). The Result sections is be split into 2 subsections. The first subsection is about predicted allosteric sites with functional residues in it. Predicted sites are ordered according to prediction confidence.

Users could click the Show Site to view the allosteric site in PDB Display Tool.
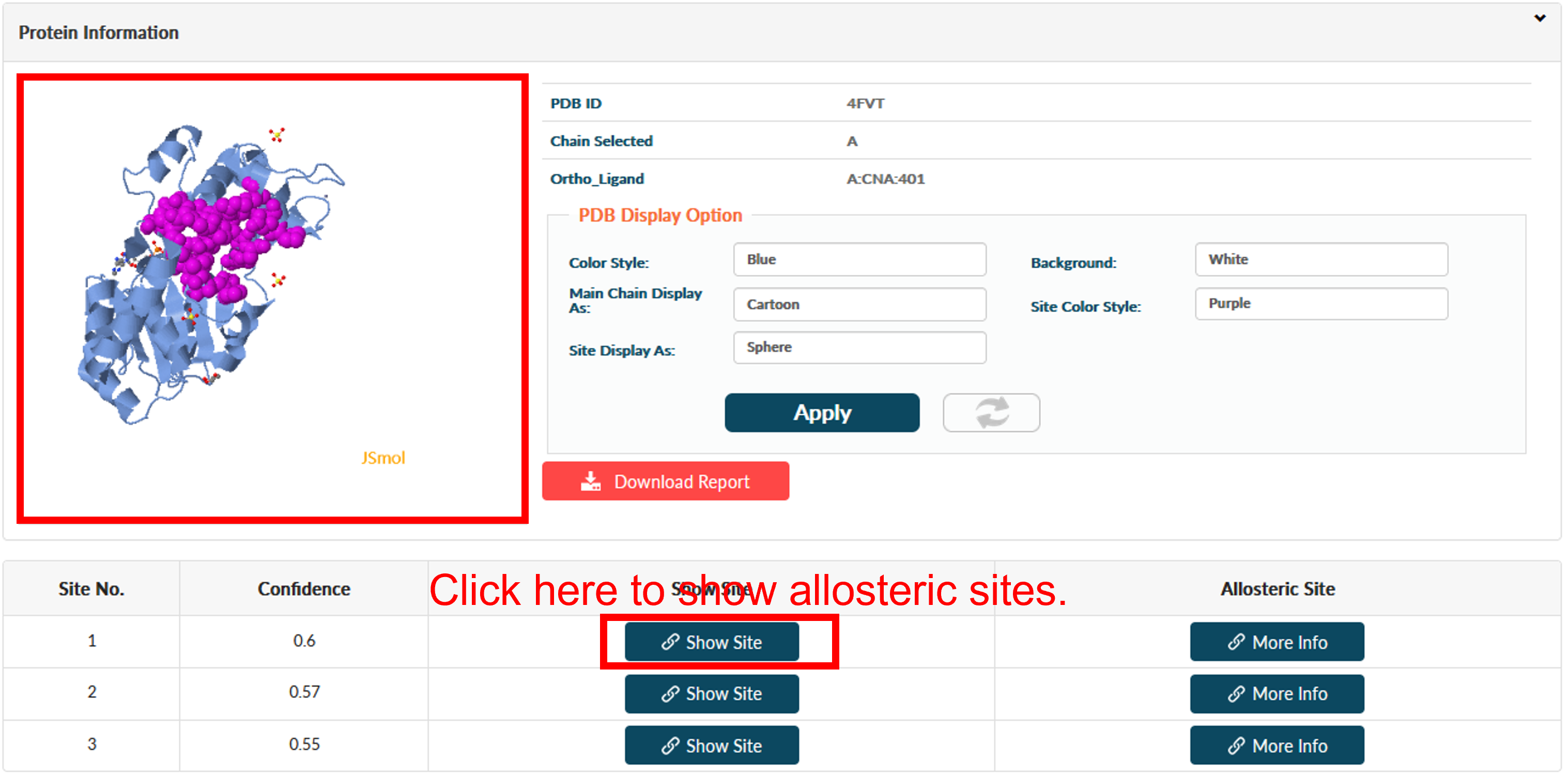
Users could click More Info for extended information about this site. Three parts of information are supported.
(1) Basic properties of the predicted pockets. This part includes volume, solvent-accessible surface area (SASA), and reversed allosteric effect (RAE) and residues in the site. Volume and SASA suggests the druggability of the pocket. RAE, or response against allosteric perturbation, is calculated by summing the RAEs of each residue in site. Site with higher RAE suggests stronger ability to regulate protein function.

(2) Key hydrophilic and hydrophobic residues. Residues in sites are plotted based on their response against orthosteric perturbation (reversed allosteric effect, RAE) and hydrophilia. Residues with high RAE are considered to have key functions. In Example 2, it could be viewed in the most confident allosteric site (Site 1), that Val 236 and Ala 252 might be important hydrophobic residues (hydrophilia < 0) while Pro 264 and Glu 266 might be vital hydrophilic residues (hydrophilia > 0). This information could help further structure-based drug design and optimization on this site.
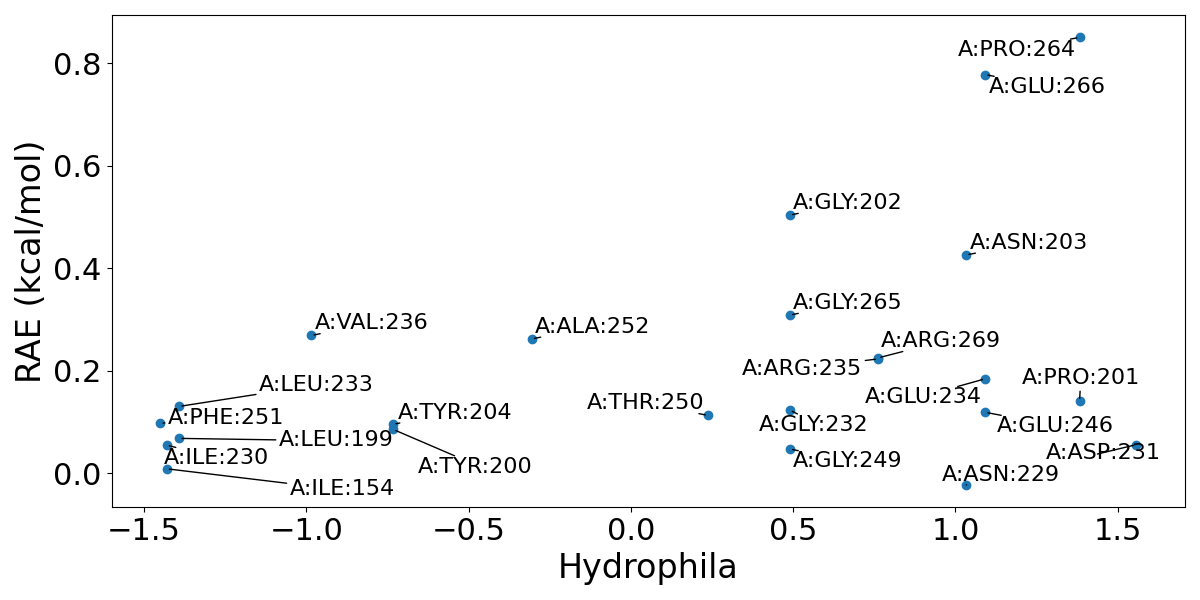
(3) Variants or Mutations. Variants and mutations recorded in Uniprot is mapped here to suggest possible functional or less functional residues in the predicted allosteric site. In Site 1 of Example 2, Asn 229 is recorded to have a mutation to Ala, and this mutation would cause the loss of function. In fact, Asn 229 is also a part of the orthosteric site and is close to orthosteric ligand. This explains its great function but with a low RAE. Note that this analysis is only available when PDB ID is used as input.

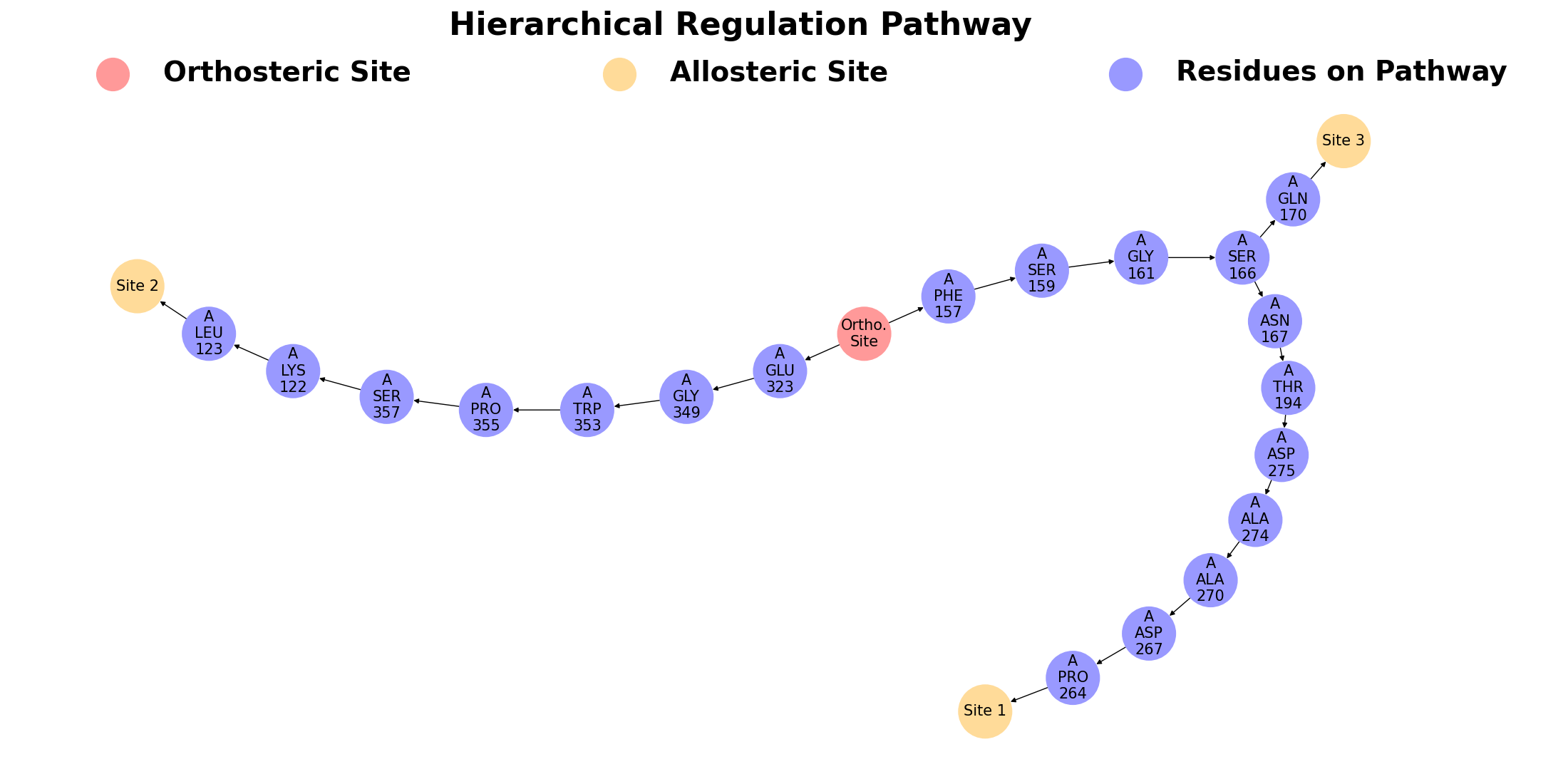
The second subsection focuses on hierarchical regulation pathways in protein, calculated from the orthosteric ligand to the most perturbated residue in site. Different allosteric regulation pathways could have couplings. A main feature of AlloReverse is to analyze the relationships among allosteric regulations and allosteric sites, offering a whole map of allostery. In SIRT3, it could be seen that the pathway towards Site 1 and 3 shares same residues (Phe 157, Ser 159, Gly 161 and Ser 166), while they have almost no relations with pathway toward Site 2.
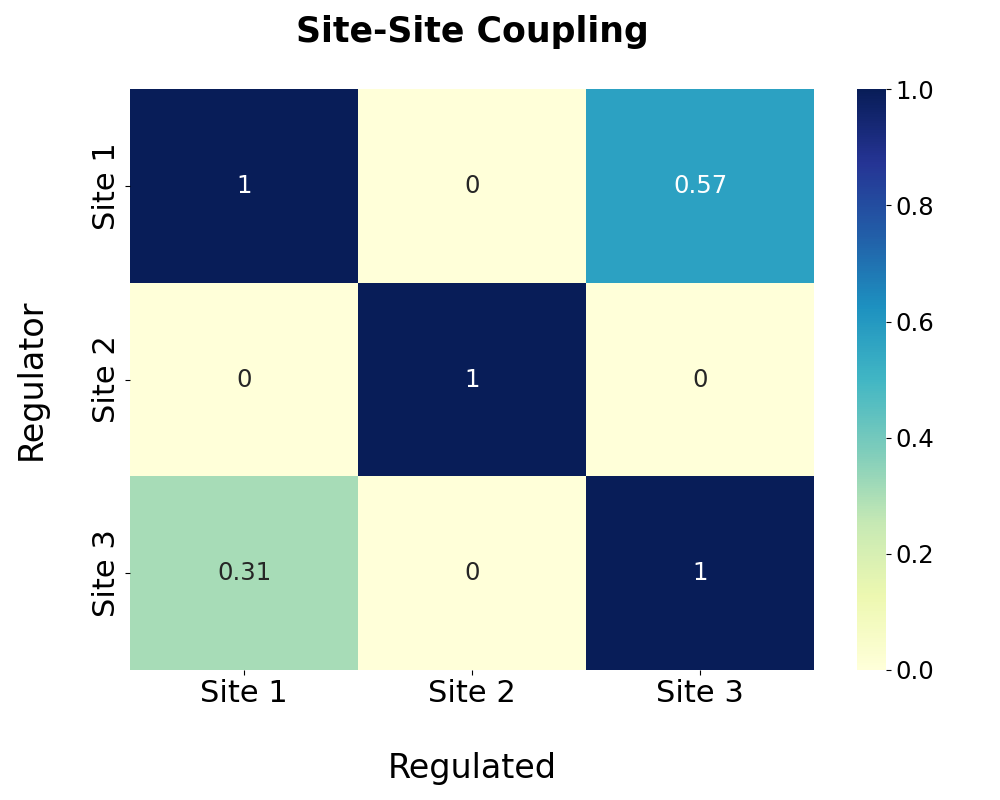
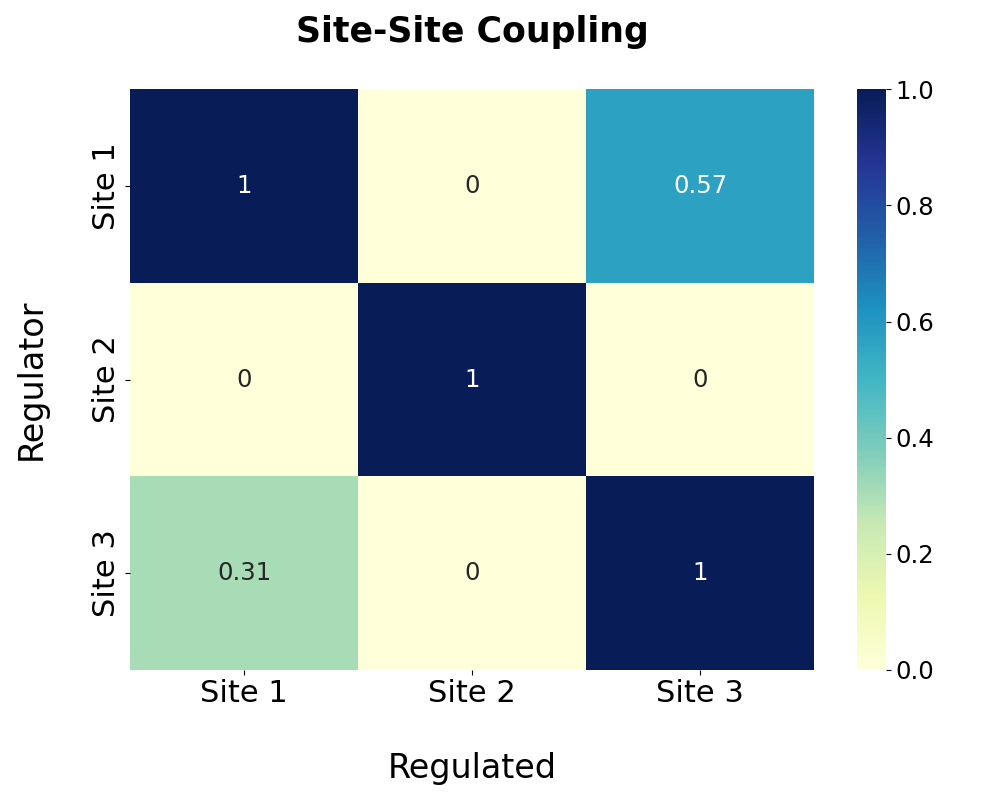
Based on the pathway analysis, regulations among predicted allosteric sites are further calculated and shown in the following heatmap. The degree of regulation of site A by site B is calculated by the proportion of pathway residues of site A shared by site B. Note that the site-site regulation analysis is only available when more than one allosteric site is predicted.
Finally, all information could be downloaded from the website. The package mainly contains a CSV containing up mentioned data and a PSE file (open by Pymol) for visualizing predicted sites.
-
Input Data Format
AlloReverse requires two inputs: Job Name and a Protein File. The Job Name could be any string. The Protein File should be an interested protein binding orthosteric (or functional) ligand. It could either be a PDB ID or a PDB format file uploaded by users.
In practice, PDB ID is more recommended. If a manual PDB file is uploaded, please pay attention to the following points.
1. Make sure the PDB file is valid. Users could check it using tools like Pymol or Discovery Studio. Invalid PDBs could not be loaded by these software.
2. Make sure that B-factors recorded in PDB files really mean B-factor. In AlloReverse analysis, B-factor is required, which is at 61st to 66th character of each record in the PDB file. In some cases, like structures from AlphaFold2, value from 61st to 66th character means prediction confidence (pLDDT) but not B-factor. This misuse might cause unrealistic predictions.
3. Orthosteric ligand is recommended to be labeled as "HETATM". Otherwise, users should input orthosteric ligand manually.
-
Output Result Explain
1. Information About Predicted Allosteric Sites
In the result page, predicted allosteric sites are ordered by their prediction confidence. The spatial region of each site could be viewed after clicking Show Site. Detailed information could be found when clicking More Info, including:
(1) Basic properties of the predicted pockets. This part includes volume, solvent-accessible surface area (SASA), reversed allosteric effect (RAE), and residues in the site. Volume and SASA suggests the druggability of the pocket. RAE, or response against allosteric perturbation, is calculated by summing the RAEs of each residue in site. Site with higher RAE suggests stronger ability to regulate protein function.

(2) Key hydrophilic and hydrophobic residues. Residues in sites are plotted based on their response against orthosteric perturbation (reversed allosteric effect, RAE) and hydrophilia. Residues with high RAE are considered to have key functions. This information could help further structure-based drug design and optimization on this site.
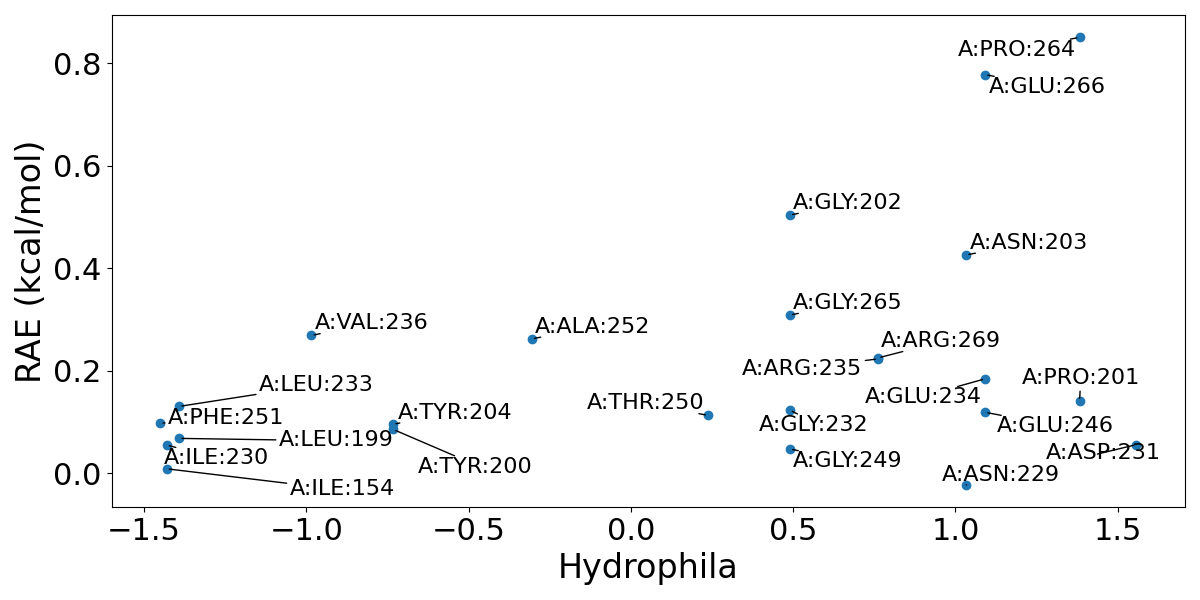
(3) Variants or Mutations. Variants and mutations recorded in Uniprot is mapped here to suggest possible functional or less functional residues in the predicted allosteric site. Usually, mutation or variants causing the loss of function is considered important, while nonsense mutations or variants are considered less important. Note that this analysis is only available when PDB ID is used as input.

2. Information About Hierarchical Allosteric Regulations
Different allosteric regulation pathways could have couplings. A main feature of AlloReverse is to analyze the relationships among allosteric regulations and allosteric sites, offering a whole map of allostery. On this subsection, the first figure shows the hierarchical allosteric regulation pathways starting from orthosteric site.
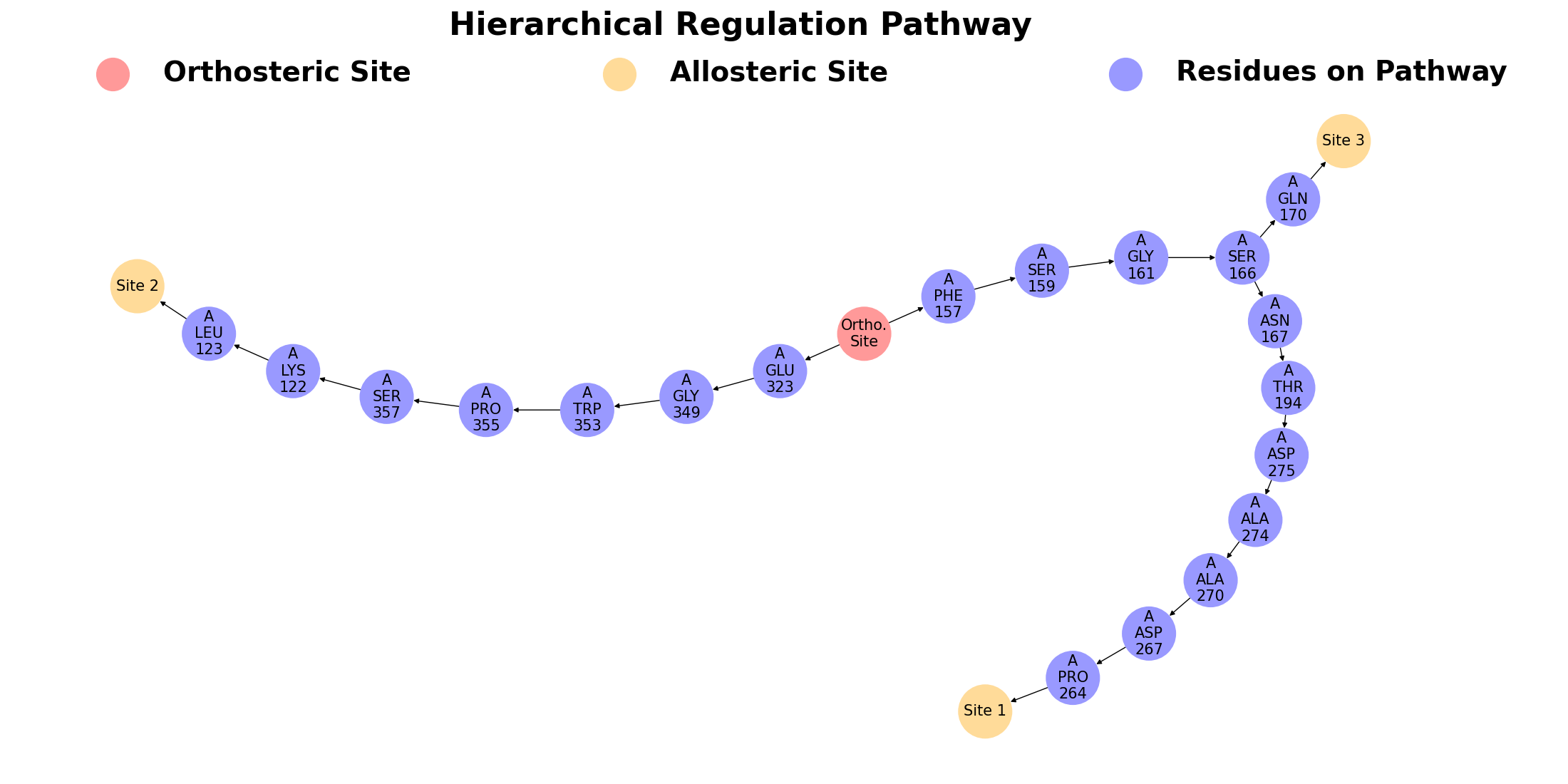
Based on the pathway analysis, regulations among predicted allosteric sites are further calculated and shown in the following heatmap. The degree of regulation of site A by site B is calculated by the proportion of pathway residues of site A shared by site B. Note that the site-site regulation analysis is only available when more than one allosteric site is predicted.
-
Job Queue
Click Job Queue to show Job Search page and here user can retrieve his/her own jobs. By inputting the type of "Job ID" and unique identifier from user in the input box, jobs of the user could be listed in a pop-up tab.